Reading All Images in a Folder Python
Why would you lot want to know more about different ways of storing and accessing images in Python? If you're segmenting a handful of images by colour or detecting faces one by ane using OpenCV, then you don't need to worry most information technology. Even if yous're using the Python Imaging Library (PIL) to draw on a few hundred photos, yous nonetheless don't need to. Storing images on deejay, as .png or .jpg files, is both suitable and appropriate.
Increasingly, however, the number of images required for a given task is getting larger and larger. Algorithms like convolutional neural networks, also known every bit convnets or CNNs, can handle enormous datasets of images and even learn from them. If yous're interested, yous can read more almost how convnets can be used for ranking selfies or for sentiment analysis.
ImageNet is a well-known public paradigm database put together for preparation models on tasks similar object nomenclature, detection, and segmentation, and information technology consists of over 14 million images.
Think almost how long it would take to load all of them into retentivity for training, in batches, maybe hundreds or thousands of times. Keep reading, and yous'll exist convinced that it would take quite awhile—at least long enough to leave your computer and do many other things while you wish y'all worked at Google or NVIDIA.
In this tutorial, you'll acquire about:
- Storing images on disk as
.pngfiles - Storing images in lightning retention-mapped databases (LMDB)
- Storing images in hierarchical data format (HDF5)
You'll likewise explore the following:
- Why alternate storage methods are worth because
- What the performance differences are when you lot're reading and writing unmarried images
- What the performance differences are when yous're reading and writing many images
- How the three methods compare in terms of disk usage
If none of the storage methods ring a bell, don't worry: for this article, all you need is a reasonably solid foundation in Python and a basic agreement of images (that they are really equanimous of multi-dimensional arrays of numbers) and relative retentiveness, such as the deviation between 10MB and 10GB.
Let'south become started!
Setup
You will need an epitome dataset to experiment with, as well equally a few Python packages.
A Dataset to Play With
We will exist using the Canadian Constitute for Avant-garde Research prototype dataset, improve known every bit CIFAR-x, which consists of sixty,000 32x32 pixel color images belonging to unlike object classes, such every bit dogs, cats, and airplanes. Relatively, CIFAR is not a very large dataset, but if we were to use the full TinyImages dataset, then you would need about 400GB of gratuitous disk infinite, which would probably exist a limiting factor.
Credits for the dataset equally described in chapter 3 of this tech report go to Alex Krizhevsky, Vinod Nair, and Geoffrey Hinton.
If you'd like to follow along with the code examples in this article, yous can download CIFAR-10 here, selecting the Python version. You'll be sacrificing 163MB of deejay space:

When yous download and unzip the binder, you'll discover that the files are not human-readable prototype files. They have actually been serialized and saved in batches using cPickle.
While we won't consider pickle or cPickle in this commodity, other than to extract the CIFAR dataset, it'due south worth mentioning that the Python pickle module has the primal advantage of existence able to serialize any Python object without whatever extra code or transformation on your function. It also has a potentially serious disadvantage of posing a security risk and not coping well when dealing with very big quantities of information.
The following code unpickles each of the 5 batch files and loads all of the images into a NumPy array:
import numpy as np import pickle from pathlib import Path # Path to the unzipped CIFAR data data_dir = Path ( "information/cifar-x-batches-py/" ) # Unpickle function provided past the CIFAR hosts def unpickle ( file ): with open ( file , "rb" ) every bit fo : dict = pickle . load ( fo , encoding = "bytes" ) return dict images , labels = [], [] for batch in data_dir . glob ( "data_batch_*" ): batch_data = unpickle ( batch ) for i , flat_im in enumerate ( batch_data [ b "information" ]): im_channels = [] # Each image is flattened, with channels in order of R, G, B for j in range ( 3 ): im_channels . append ( flat_im [ j * 1024 : ( j + 1 ) * 1024 ] . reshape (( 32 , 32 )) ) # Reconstruct the original epitome images . suspend ( np . dstack (( im_channels ))) # Save the characterization labels . append ( batch_data [ b "labels" ][ i ]) impress ( "Loaded CIFAR-10 preparation prepare:" ) print ( f " - np.shape(images) { np . shape ( images ) } " ) print ( f " - np.shape(labels) { np . shape ( labels ) } " ) All the images are now in RAM in the images variable, with their corresponding meta information in labels, and are ready for you to manipulate. Adjacent, you can install the Python packages you'll use for the three methods.
Setup for Storing Images on Disk
Yous'll need to fix your environment for the default method of saving and accessing these images from disk. This article will assume y'all have Python 3.x installed on your system, and volition use Pillow for the image manipulation:
Alternatively, if you prefer, you tin can install information technology using Anaconda:
$ conda install -c conda-forge pillow Now you lot're ready for storing and reading images from disk.
Getting Started With LMDB
LMDB, sometimes referred to as the "Lightning Database," stands for Lightning Memory-Mapped Database because it'due south fast and uses memory-mapped files. It's a fundamental-value store, non a relational database.
In terms of implementation, LMDB is a B+ tree, which basically ways that it is a tree-like graph construction stored in memory where each fundamental-value element is a node, and nodes can have many children. Nodes on the same level are linked to one another for fast traversal.
Critically, key components of the B+ tree are set to stand for to the page size of the host operating system, maximizing efficiency when accessing any key-value pair in the database. Since LMDB high-performance heavily relies on this particular point, LMDB efficiency has been shown to exist dependent on the underlying file system and its implementation.
Some other key reason for the efficiency of LMDB is that it is retention-mapped. This means that information technology returns direct pointers to the memory addresses of both keys and values, without needing to copy annihilation in memory as most other databases do.
Those who want to dive into a scrap more of the internal implementation details of B+ trees can check out this article on B+ trees and so play with this visualization of node insertion.
If B+ trees don't involvement you, don't worry. You don't need to know much about their internal implementation in order to use LMDB. Nosotros will be using the Python binding for the LMDB C library, which tin exist installed via pip:
You too have the option of installing via Anaconda:
$ conda install -c conda-forge python-lmdb Check that y'all tin can import lmdb from a Python crush, and you're good to become.
Getting Started With HDF5
HDF5 stands for Hierarchical Data Format, a file format referred to equally HDF4 or HDF5. We don't need to worry about HDF4, every bit HDF5 is the current maintained version.
Interestingly, HDF has its origins in the National Eye for Supercomputing Applications, as a portable, compact scientific data format. If you lot're wondering if it's widely used, check out NASA's blurb on HDF5 from their Earth Information project.
HDF files consist of two types of objects:
- Datasets
- Groups
Datasets are multidimensional arrays, and groups consist of datasets or other groups. Multidimensional arrays of whatsoever size and type can be stored as a dataset, but the dimensions and type take to be uniform within a dataset. Each dataset must contain a homogeneous N-dimensional array. That said, because groups and datasets may be nested, you can still get the heterogeneity you lot may need:
Every bit with the other libraries, you can alternately install via Anaconda:
$ conda install -c conda-forge h5py If you can import h5py from a Python vanquish, everything is set up properly.
Storing a Single Paradigm
Now that you have a full general overview of the methods, permit's dive straight in and expect at a quantitative comparison of the basic tasks nosotros care about: how long it takes to read and write files, and how much disk memory will be used. This will also serve as a basic introduction to how the methods work, with lawmaking examples of how to use them.
When I refer to "files," I generally mean a lot of them. However, it is important to make a distinction since some methods may exist optimized for different operations and quantities of files.
For the purposes of experimentation, we tin compare the performance between various quantities of files, by factors of 10 from a single image to 100,000 images. Since our 5 batches of CIFAR-10 add up to 50,000 images, nosotros tin can use each image twice to go to 100,000 images.
To prepare for the experiments, you will want to create a binder for each method, which volition comprise all the database files or images, and salve the paths to those directories in variables:
from pathlib import Path disk_dir = Path ( "data/disk/" ) lmdb_dir = Path ( "data/lmdb/" ) hdf5_dir = Path ( "data/hdf5/" ) Path does not automatically create the folders for you lot unless y'all specifically enquire it to:
disk_dir . mkdir ( parents = True , exist_ok = Truthful ) lmdb_dir . mkdir ( parents = True , exist_ok = True ) hdf5_dir . mkdir ( parents = Truthful , exist_ok = True ) Now y'all can move on to running the actual experiments, with lawmaking examples of how to perform bones tasks with the three different methods. We can use the timeit module, which is included in the Python standard library, to help time the experiments.
Although the master purpose of this commodity is not to larn the APIs of the unlike Python packages, it is helpful to take an understanding of how they can exist implemented. We will go through the full general principles alongside all the code used to behave the storing experiments.
Storing to Disk
Our input for this experiment is a unmarried image paradigm, currently in memory every bit a NumPy array. Yous want to save information technology commencement to disk equally a .png paradigm, and name it using a unique paradigm ID image_id. This can exist done using the Pillow parcel you installed earlier:
from PIL import Image import csv def store_single_disk ( image , image_id , label ): """ Stores a single image every bit a .png file on disk. Parameters: --------------- image image array, (32, 32, iii) to be stored image_id integer unique ID for image label epitome label """ Image . fromarray ( image ) . salve ( disk_dir / f " { image_id } .png" ) with open ( disk_dir / f " { image_id } .csv" , "wt" ) as csvfile : writer = csv . writer ( csvfile , delimiter = " " , quotechar = "|" , quoting = csv . QUOTE_MINIMAL ) writer . writerow ([ label ]) This saves the image. In all realistic applications, you also care about the meta data fastened to the image, which in our example dataset is the image label. When y'all're storing images to disk, there are several options for saving the meta data.
I solution is to encode the labels into the image name. This has the reward of not requiring whatever extra files.
However, it besides has the large disadvantage of forcing you to bargain with all the files whenever you practise anything with labels. Storing the labels in a dissever file allows y'all to play around with the labels alone, without having to load the images. Above, I accept stored the labels in a separate .csv files for this experiment.
Now allow's move on to doing the exact same task with LMDB.
Storing to LMDB
Firstly, LMDB is a key-value storage system where each entry is saved every bit a byte assortment, so in our case, keys will be a unique identifier for each image, and the value will exist the prototype itself. Both the keys and values are expected to be strings, so the common usage is to serialize the value as a cord, and and then unserialize it when reading it back out.
Yous tin apply pickle for the serializing. Whatever Python object can exist serialized, so yous might equally well include the image meta data in the database as well. This saves you lot the trouble of attaching meta data dorsum to the image data when nosotros load the dataset from disk.
You tin can create a basic Python class for the image and its meta data:
class CIFAR_Image : def __init__ ( self , image , characterization ): # Dimensions of image for reconstruction - non really necessary # for this dataset, but some datasets may include images of # varying sizes self . channels = prototype . shape [ 2 ] self . size = image . shape [: ii ] self . epitome = epitome . tobytes () self . label = label def get_image ( self ): """ Returns the image equally a numpy array. """ prototype = np . frombuffer ( self . epitome , dtype = np . uint8 ) return image . reshape ( * cocky . size , cocky . channels ) Secondly, because LMDB is memory-mapped, new databases need to know how much memory they are expected to apply up. This is relatively straightforward in our case, but it tin exist a massive pain in other cases, which you will see in more depth in a subsequently section. LMDB calls this variable the map_size.
Finally, read and write operations with LMDB are performed in transactions. You can think of them equally like to those of a traditional database, consisting of a group of operations on the database. This may wait already significantly more complicated than the disk version, but hang on and keep reading!
With those three points in mind, let'south look at the code to save a single image to a LMDB:
import lmdb import pickle def store_single_lmdb ( image , image_id , label ): """ Stores a unmarried image to a LMDB. Parameters: --------------- epitome prototype array, (32, 32, 3) to be stored image_id integer unique ID for image label paradigm label """ map_size = image . nbytes * 10 # Create a new LMDB surroundings env = lmdb . open ( str ( lmdb_dir / f "single_lmdb" ), map_size = map_size ) # Start a new write transaction with env . begin ( write = Truthful ) as txn : # All key-value pairs need to exist strings value = CIFAR_Image ( image , label ) key = f " { image_id : 08 } " txn . put ( key . encode ( "ascii" ), pickle . dumps ( value )) env . close () Yous are now set up to save an image to LMDB. Lastly, let's look at the final method, HDF5.
Storing With HDF5
Remember that an HDF5 file can contain more than than one dataset. In this rather picayune case, you tin can create 2 datasets, one for the image, and one for its meta data:
import h5py def store_single_hdf5 ( image , image_id , label ): """ Stores a single epitome to an HDF5 file. Parameters: --------------- image image array, (32, 32, iii) to exist stored image_id integer unique ID for image label prototype characterization """ # Create a new HDF5 file file = h5py . File ( hdf5_dir / f " { image_id } .h5" , "w" ) # Create a dataset in the file dataset = file . create_dataset ( "image" , np . shape ( image ), h5py . h5t . STD_U8BE , data = image ) meta_set = file . create_dataset ( "meta" , np . shape ( label ), h5py . h5t . STD_U8BE , information = label ) file . shut () h5py.h5t.STD_U8BE specifies the blazon of data that will exist stored in the dataset, which in this case is unsigned 8-scrap integers. You can see a full list of HDF'due south predefined datatypes here.
Now that we have reviewed the three methods of saving a single image, let'southward motility on to the next footstep.
Experiments for Storing a Single Image
Now yous tin can put all iii functions for saving a single image into a dictionary, which can be called afterwards during the timing experiments:
_store_single_funcs = dict ( disk = store_single_disk , lmdb = store_single_lmdb , hdf5 = store_single_hdf5 ) Finally, everything is gear up for conducting the timed experiment. Let'south attempt saving the showtime prototype from CIFAR and its respective label, and storing it in the iii different means:
from timeit import timeit store_single_timings = dict () for method in ( "deejay" , "lmdb" , "hdf5" ): t = timeit ( "_store_single_funcs[method](paradigm, 0, label)" , setup = "prototype=images[0]; label=labels[0]" , number = 1 , globals = globals (), ) store_single_timings [ method ] = t print ( f "Method: { method } , Time usage: { t } " ) Remember that we're interested in runtime, displayed hither in seconds, and also the retentiveness usage:
| Method | Save Single Paradigm + Meta | Memory |
|---|---|---|
| Deejay | 1.915 ms | 8 One thousand |
| LMDB | 1.203 ms | 32 K |
| HDF5 | 8.243 ms | 8 One thousand |
There are two takeaways here:
- All of the methods are trivially quick.
- In terms of disk usage, LMDB uses more than.
Clearly, despite LMDB having a slight performance lead, we haven't convinced anyone why to not just store images on deejay. After all, it's a homo readable format, and y'all tin open up and view them from any file system browser! Well, it's time to expect at a lot more images…
Storing Many Images
You have seen the code for using the various storage methods to salvage a single image, so at present we need to adjust the lawmaking to relieve many images and and so run the timed experiment.
Adjusting the Code for Many Images
Saving multiple images every bit .png files is as straightforward as calling store_single_method() multiple times. But this isn't truthful for LMDB or HDF5, since you don't desire a different database file for each image. Rather, you want to put all of the images into one or more than files.
You will need to slightly alter the code and create three new functions that accept multiple images, store_many_disk(), store_many_lmdb(), and store_many_hdf5:
store_many_disk ( images , labels ): """ Stores an assortment of images to disk Parameters: --------------- images images array, (N, 32, 32, 3) to be stored labels labels array, (N, 1) to be stored """ num_images = len ( images ) # Save all the images one by one for i , prototype in enumerate ( images ): Paradigm . fromarray ( prototype ) . relieve ( disk_dir / f " { i } .png" ) # Save all the labels to the csv file with open ( disk_dir / f " { num_images } .csv" , "w" ) as csvfile : writer = csv . writer ( csvfile , delimiter = " " , quotechar = "|" , quoting = csv . QUOTE_MINIMAL ) for characterization in labels : # This typically would exist more than only one value per row writer . writerow ([ label ]) def store_many_lmdb ( images , labels ): """ Stores an array of images to LMDB. Parameters: --------------- images images array, (North, 32, 32, three) to be stored labels labels array, (N, 1) to be stored """ num_images = len ( images ) map_size = num_images * images [ 0 ] . nbytes * 10 # Create a new LMDB DB for all the images env = lmdb . open ( str ( lmdb_dir / f " { num_images } _lmdb" ), map_size = map_size ) # Aforementioned as before — merely allow's write all the images in a single transaction with env . begin ( write = True ) as txn : for i in range ( num_images ): # All fundamental-value pairs need to be Strings value = CIFAR_Image ( images [ i ], labels [ i ]) central = f " { i : 08 } " txn . put ( key . encode ( "ascii" ), pickle . dumps ( value )) env . close () def store_many_hdf5 ( images , labels ): """ Stores an assortment of images to HDF5. Parameters: --------------- images images assortment, (Due north, 32, 32, 3) to be stored labels labels array, (N, ane) to exist stored """ num_images = len ( images ) # Create a new HDF5 file file = h5py . File ( hdf5_dir / f " { num_images } _many.h5" , "due west" ) # Create a dataset in the file dataset = file . create_dataset ( "images" , np . shape ( images ), h5py . h5t . STD_U8BE , information = images ) meta_set = file . create_dataset ( "meta" , np . shape ( labels ), h5py . h5t . STD_U8BE , information = labels ) file . close () So you could store more one file to deejay, the image files method was altered to loop over each image in the list. For LMDB, a loop is likewise needed since we are creating a CIFAR_Image object for each image and its meta data.
The smallest adjustment is with the HDF5 method. In fact, at that place's hardly an adjustment at all! HFD5 files accept no limitation on file size aside from external restrictions or dataset size, and then all the images were stuffed into a unmarried dataset, just like before.
Next, you volition need to prepare the dataset for the experiments past increasing its size.
Preparing the Dataset
Before running the experiments again, allow's first double our dataset size so that we can exam with up to 100,000 images:
cutoffs = [ ten , 100 , thousand , 10000 , 100000 ] # Let'southward double our images so that nosotros accept 100,000 images = np . concatenate (( images , images ), axis = 0 ) labels = np . concatenate (( labels , labels ), axis = 0 ) # Make sure you actually take 100,000 images and labels print ( np . shape ( images )) print ( np . shape ( labels )) At present that in that location are plenty images, it'south time for the experiment.
Experiment for Storing Many Images
As you did with reading many images, you can create a dictionary handling all the functions with store_many_ and run the experiments:
_store_many_funcs = dict ( disk = store_many_disk , lmdb = store_many_lmdb , hdf5 = store_many_hdf5 ) from timeit import timeit store_many_timings = { "disk" : [], "lmdb" : [], "hdf5" : []} for cutoff in cutoffs : for method in ( "disk" , "lmdb" , "hdf5" ): t = timeit ( "_store_many_funcs[method](images_, labels_)" , setup = "images_=images[:cutoff]; labels_=labels[:cutoff]" , number = one , globals = globals (), ) store_many_timings [ method ] . append ( t ) # Print out the method, cutoff, and elapsed time print ( f "Method: { method } , Time usage: { t } " ) If you're following along and running the code yourself, you'll need to sit down dorsum a moment in suspense and wait for 111,110 images to exist stored three times each to your disk, in three dissimilar formats. You'll besides need to say goodbye to approximately 2 GB of disk space.
Now for the moment of truth! How long did all of that storing take? A picture is worth a 1000 words:

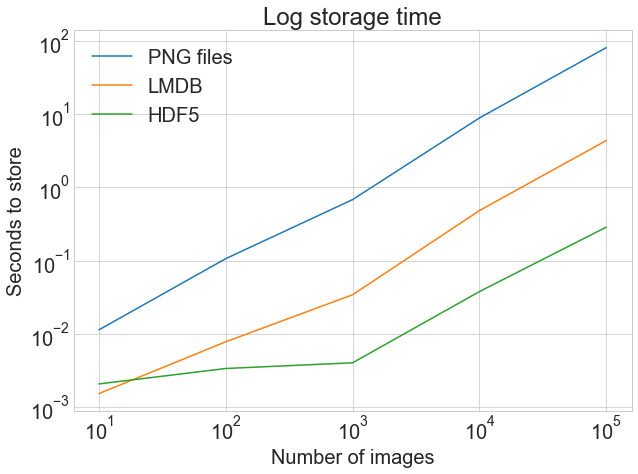
The first graph shows the normal, unadjusted storage time, highlighting the drastic difference between storing to .png files and LMDB or HDF5.
The 2d graph shows the log of the timings, highlighting that HDF5 starts out slower than LMDB but, with larger quantities of images, comes out slightly alee.
While exact results may vary depending on your motorcar, this is why LMDB and HDF5 are worth thinking about. Here'southward the code that generated the above graph:
import matplotlib.pyplot as plt def plot_with_legend ( x_range , y_data , legend_labels , x_label , y_label , title , log = False ): """ Displays a single plot with multiple datasets and matching legends. Parameters: -------------- x_range list of lists containing x information y_data listing of lists containing y values legend_labels list of cord legend labels x_label x axis label y_label y axis characterization """ plt . style . use ( "seaborn-whitegrid" ) plt . figure ( figsize = ( 10 , seven )) if len ( y_data ) != len ( legend_labels ): raise TypeError ( "Error: number of data sets does non match number of labels." ) all_plots = [] for information , label in nada ( y_data , legend_labels ): if log : temp , = plt . loglog ( x_range , data , characterization = label ) else : temp , = plt . plot ( x_range , information , label = characterization ) all_plots . append ( temp ) plt . title ( title ) plt . xlabel ( x_label ) plt . ylabel ( y_label ) plt . legend ( handles = all_plots ) plt . bear witness () # Getting the store timings data to brandish disk_x = store_many_timings [ "disk" ] lmdb_x = store_many_timings [ "lmdb" ] hdf5_x = store_many_timings [ "hdf5" ] plot_with_legend ( cutoffs , [ disk_x , lmdb_x , hdf5_x ], [ "PNG files" , "LMDB" , "HDF5" ], "Number of images" , "Seconds to store" , "Storage time" , log = Faux , ) plot_with_legend ( cutoffs , [ disk_x , lmdb_x , hdf5_x ], [ "PNG files" , "LMDB" , "HDF5" ], "Number of images" , "Seconds to store" , "Log storage fourth dimension" , log = True , ) At present let'due south continue to reading the images back out.
Reading a Single Paradigm
First, let's consider the case for reading a single image back into an array for each of the three methods.
Reading From Disk
Of the three methods, LMDB requires the most legwork when reading image files dorsum out of memory, because of the serialization step. Let's walk through these functions that read a single image out for each of the three storage formats.
First, read a unmarried prototype and its meta from a .png and .csv file:
def read_single_disk ( image_id ): """ Stores a unmarried image to disk. Parameters: --------------- image_id integer unique ID for image Returns: ---------- prototype prototype array, (32, 32, 3) to exist stored label associated meta data, int label """ paradigm = np . array ( Prototype . open ( disk_dir / f " { image_id } .png" )) with open up ( disk_dir / f " { image_id } .csv" , "r" ) equally csvfile : reader = csv . reader ( csvfile , delimiter = " " , quotechar = "|" , quoting = csv . QUOTE_MINIMAL ) label = int ( next ( reader )[ 0 ]) render epitome , label Reading From LMDB
Next, read the same image and meta from an LMDB by opening the surroundings and starting a read transaction:
1 def read_single_lmdb ( image_id ): two """ Stores a single image to LMDB. iii Parameters: iv --------------- 5 image_id integer unique ID for image 6 7 Returns: viii ---------- 9 image image assortment, (32, 32, 3) to exist stored ten characterization associated meta information, int label 11 """ 12 # Open up the LMDB environs xiii env = lmdb . open ( str ( lmdb_dir / f "single_lmdb" ), readonly = True ) xiv 15 # Start a new read transaction 16 with env . begin () every bit txn : 17 # Encode the central the aforementioned way as we stored it eighteen data = txn . get ( f " { image_id : 08 } " . encode ( "ascii" )) 19 # Remember information technology's a CIFAR_Image object that is loaded xx cifar_image = pickle . loads ( data ) 21 # Retrieve the relevant $.25 22 epitome = cifar_image . get_image () 23 label = cifar_image . label 24 env . shut () 25 26 return image , label Here are a couple points to not about the lawmaking snippet above:
- Line 13: The
readonly=Trueflag specifies that no writes will be allowed on the LMDB file until the transaction is finished. In database lingo, information technology's equivalent to taking a read lock. - Line xx: To recollect the CIFAR_Image object, you demand to contrary the steps we took to pickle information technology when nosotros were writing it. This is where the
get_image()of the object is helpful.
This wraps up reading the image dorsum out from LMDB. Finally, you volition desire to do the aforementioned with HDF5.
Reading From HDF5
Reading from HDF5 looks very similar to the writing procedure. Here is the code to open and read the HDF5 file and parse the same prototype and meta:
def read_single_hdf5 ( image_id ): """ Stores a single image to HDF5. Parameters: --------------- image_id integer unique ID for image Returns: ---------- paradigm epitome array, (32, 32, iii) to be stored characterization associated meta information, int characterization """ # Open the HDF5 file file = h5py . File ( hdf5_dir / f " { image_id } .h5" , "r+" ) prototype = np . assortment ( file [ "/image" ]) . astype ( "uint8" ) label = int ( np . array ( file [ "/meta" ]) . astype ( "uint8" )) return image , label Note that you lot access the various datasets in the file by indexing the file object using the dataset name preceded by a forward slash /. As earlier, y'all tin can create a dictionary containing all the read functions:
_read_single_funcs = dict ( disk = read_single_disk , lmdb = read_single_lmdb , hdf5 = read_single_hdf5 ) With this dictionary prepared, you are ready for running the experiment.
Experiment for Reading a Single Paradigm
Yous might expect that the experiment for reading a single prototype in will accept somewhat trivial results, only here's the experiment code:
from timeit import timeit read_single_timings = dict () for method in ( "disk" , "lmdb" , "hdf5" ): t = timeit ( "_read_single_funcs[method](0)" , setup = "paradigm=images[0]; label=labels[0]" , number = 1 , globals = globals (), ) read_single_timings [ method ] = t print ( f "Method: { method } , Time usage: { t } " ) Here are the results of the experiment for reading a single prototype:
| Method | Read Unmarried Image + Meta |
|---|---|
| Deejay | 1.61970 ms |
| LMDB | iv.52063 ms |
| HDF5 | one.98036 ms |
It's slightly faster to read the .png and .csv files directly from deejay, simply all three methods perform trivially speedily. The experiments we'll practice side by side are much more interesting.
Reading Many Images
Now you can accommodate the code to read many images at one time. This is probable the action you'll be performing most ofttimes, so the runtime performance is essential.
Adjusting the Lawmaking for Many Images
Extending the functions above, you can create functions with read_many_, which can exist used for the next experiments. Like before, it is interesting to compare performance when reading different quantities of images, which are repeated in the code below for reference:
def read_many_disk ( num_images ): """ Reads paradigm from deejay. Parameters: --------------- num_images number of images to read Returns: ---------- images images array, (N, 32, 32, three) to be stored labels associated meta data, int label (N, 1) """ images , labels = [], [] # Loop over all IDs and read each image in one by one for image_id in range ( num_images ): images . suspend ( np . array ( Epitome . open ( disk_dir / f " { image_id } .png" ))) with open up ( disk_dir / f " { num_images } .csv" , "r" ) as csvfile : reader = csv . reader ( csvfile , delimiter = " " , quotechar = "|" , quoting = csv . QUOTE_MINIMAL ) for row in reader : labels . append ( int ( row [ 0 ])) return images , labels def read_many_lmdb ( num_images ): """ Reads image from LMDB. Parameters: --------------- num_images number of images to read Returns: ---------- images images array, (N, 32, 32, 3) to exist stored labels associated meta data, int label (Due north, i) """ images , labels = [], [] env = lmdb . open up ( str ( lmdb_dir / f " { num_images } _lmdb" ), readonly = True ) # Start a new read transaction with env . begin () as txn : # Read all images in one single transaction, with one lock # Nosotros could carve up this up into multiple transactions if needed for image_id in range ( num_images ): data = txn . get ( f " { image_id : 08 } " . encode ( "ascii" )) # Remember that information technology'south a CIFAR_Image object # that is stored every bit the value cifar_image = pickle . loads ( information ) # Think the relevant bits images . append ( cifar_image . get_image ()) labels . append ( cifar_image . label ) env . shut () return images , labels def read_many_hdf5 ( num_images ): """ Reads prototype from HDF5. Parameters: --------------- num_images number of images to read Returns: ---------- images images array, (Northward, 32, 32, iii) to be stored labels associated meta information, int characterization (North, 1) """ images , labels = [], [] # Open up the HDF5 file file = h5py . File ( hdf5_dir / f " { num_images } _many.h5" , "r+" ) images = np . array ( file [ "/images" ]) . astype ( "uint8" ) labels = np . assortment ( file [ "/meta" ]) . astype ( "uint8" ) return images , labels _read_many_funcs = dict ( disk = read_many_disk , lmdb = read_many_lmdb , hdf5 = read_many_hdf5 ) With the reading functions stored in a dictionary every bit with the writing functions, you lot're all set for the experiment.
Experiment for Reading Many Images
You can now run the experiment for reading many images out:
from timeit import timeit read_many_timings = { "disk" : [], "lmdb" : [], "hdf5" : []} for cutoff in cutoffs : for method in ( "deejay" , "lmdb" , "hdf5" ): t = timeit ( "_read_many_funcs[method](num_images)" , setup = "num_images=cutoff" , number = one , globals = globals (), ) read_many_timings [ method ] . suspend ( t ) # Impress out the method, cutoff, and elapsed fourth dimension print ( f "Method: { method } , No. images: { cutoff } , Time usage: { t } " ) As we did previously, you can graph the read experiment results:


The top graph shows the normal, unadjusted read times, showing the drastic departure between reading from .png files and LMDB or HDF5.
In dissimilarity, the graph on the lesser shows the log of the timings, highlighting the relative differences with fewer images. Namely, we can see how HDF5 starts out backside simply, with more images, becomes consistently faster than LMDB by a small margin.
Using the aforementioned plotting function as for the write timings, we have the post-obit:
disk_x_r = read_many_timings [ "disk" ] lmdb_x_r = read_many_timings [ "lmdb" ] hdf5_x_r = read_many_timings [ "hdf5" ] plot_with_legend ( cutoffs , [ disk_x_r , lmdb_x_r , hdf5_x_r ], [ "PNG files" , "LMDB" , "HDF5" ], "Number of images" , "Seconds to read" , "Read time" , log = False , ) plot_with_legend ( cutoffs , [ disk_x_r , lmdb_x_r , hdf5_x_r ], [ "PNG files" , "LMDB" , "HDF5" ], "Number of images" , "Seconds to read" , "Log read fourth dimension" , log = True , ) In practice, the write time is ofttimes less disquisitional than the read time. Imagine that you are training a deep neural network on images, and just half of your unabridged image dataset fits into RAM at in one case. Each epoch of training a network requires the unabridged dataset, and the model needs a few hundred epochs to converge. You will substantially be reading half of the dataset into retention every epoch.
There are several tricks people do, such as training pseudo-epochs to brand this slightly amend, but you go the idea.
At present, expect again at the read graph above. The difference between a 40-2d and four-second read time suddenly is the departure between waiting half-dozen hours for your model to railroad train, or forty minutes!
If we view the read and write times on the same chart, we accept the following:
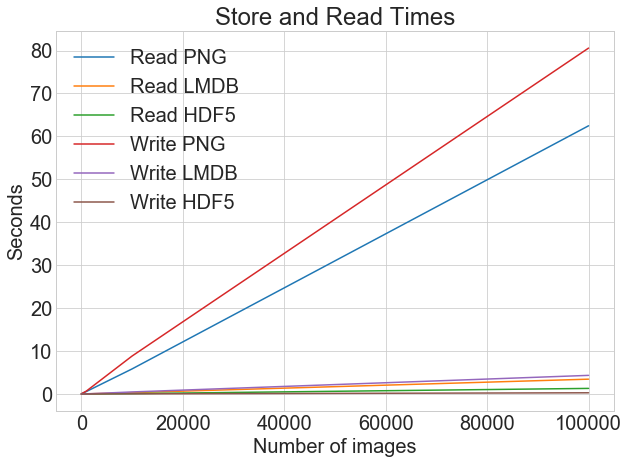
You can plot all the read and write timings on a unmarried graph using the same plotting function:
plot_with_legend ( cutoffs , [ disk_x_r , lmdb_x_r , hdf5_x_r , disk_x , lmdb_x , hdf5_x ], [ "Read PNG" , "Read LMDB" , "Read HDF5" , "Write PNG" , "Write LMDB" , "Write HDF5" , ], "Number of images" , "Seconds" , "Log Store and Read Times" , log = False , ) When y'all're storing images equally .png files, there is a large difference between write and read times. Nevertheless, with LMDB and HDF5, the divergence is much less marked. Overall, even if read fourth dimension is more critical than write time, at that place is a strong argument for storing images using LMDB or HDF5.
At present that you've seen the performance benefits of LMDB and HDF5, let'south look at another crucial metric: deejay usage.
Because Deejay Usage
Speed is not the merely performance metric you may be interested in. Nosotros're already dealing with very large datasets, and so deejay space is as well a very valid and relevant concern.
Suppose you have an image dataset of 3TB. Presumably, you have them already on disk somewhere, dissimilar our CIFAR example, and then by using an alternate storage method, you lot are essentially making a copy of them, which also has to be stored. Doing and then will give you huge performance benefits when yous use the images, simply you'll demand to make certain you lot have enough disk space.
How much disk space practise the various storage methods apply? Here's the disk space used for each method for each quantity of images:
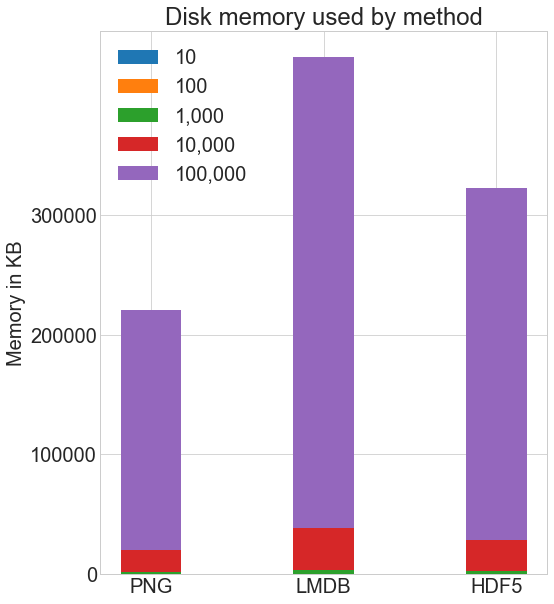
I used the Linux du -h -c folder_name/* command to compute the disk usage on my organisation. There is some approximation inherent with this method due to rounding, but here's the general comparison:
# Memory used in KB disk_mem = [ 24 , 204 , 2004 , 20032 , 200296 ] lmdb_mem = [ lx , 420 , 4000 , 39000 , 393000 ] hdf5_mem = [ 36 , 304 , 2900 , 29000 , 293000 ] X = [ disk_mem , lmdb_mem , hdf5_mem ] ind = np . arange ( iii ) width = 0.35 plt . subplots ( figsize = ( 8 , 10 )) plots = [ plt . bar ( ind , [ row [ 0 ] for row in 10 ], width )] for i in range ( i , len ( cutoffs )): plots . append ( plt . bar ( ind , [ row [ i ] for row in X ], width , bottom = [ row [ i - i ] for row in X ] ) ) plt . ylabel ( "Memory in KB" ) plt . title ( "Disk memory used past method" ) plt . xticks ( ind , ( "PNG" , "LMDB" , "HDF5" )) plt . yticks ( np . arange ( 0 , 400000 , 100000 )) plt . legend ( [ plot [ 0 ] for plot in plots ], ( "10" , "100" , "1,000" , "x,000" , "100,000" ) ) plt . show () Both HDF5 and LMDB have upward more disk infinite than if you store using normal .png images. It's important to annotation that both LMDB and HDF5 disk usage and performance depend highly on diverse factors, including operating arrangement and, more critically, the size of the data you lot store.
LMDB gains its efficiency from caching and taking advantage of Os page sizes. You don't demand to understand its inner workings, only notation that with larger images, you will end up with significantly more than disk usage with LMDB, because images won't fit on LMDB'south leaf pages, the regular storage location in the tree, and instead y'all will have many overflow pages. The LMDB bar in the chart higher up will shoot off the chart.
Our 32x32x3 pixel images are relatively modest compared to the average images you may use, and they let for optimal LMDB performance.
While we won't explore it hither experimentally, in my own experience with images of 256x256x3 or 512x512x3 pixels, HDF5 is commonly slightly more efficient in terms of disk usage than LMDB. This is a adept transition into the final department, a qualitative discussion of the differences betwixt the methods.
Word
There are other distinguishing features of LMDB and HDF5 that are worth knowing about, and it's likewise important to briefly hash out some of the criticisms of both methods. Several links are included along with the word if you want to learn more.
Parallel Access
A fundamental comparison that we didn't examination in the experiments to a higher place is concurrent reads and writes. Often, with such big datasets, you may want to speed upwardly your operation through parallelization.
In the bulk of cases, you won't exist interested in reading parts of the same image at the same time, but you will want to read multiple images at once. With this definition of concurrency, storing to disk equally .png files actually allows for consummate concurrency. Zip prevents you from reading several images at one time from different threads, or writing multiple files at once, equally long equally the image names are different.
How about LMDB? There can exist multiple readers on an LMDB environs at a fourth dimension, but only i writer, and writers practice not block readers. You lot can read more about that at the LMDB technology website.
Multiple applications can access the aforementioned LMDB database at the aforementioned time, and multiple threads from the aforementioned process can too meantime admission the LMDB for reads. This allows for fifty-fifty quicker read times: if you divided all of CIFAR into ten sets, then you lot could ready ten processes to each read in i set up, and it would divide the loading fourth dimension by ten.
HDF5 also offers parallel I/O, allowing concurrent reads and writes. However, in implementation, a write lock is held, and access is sequential, unless you have a parallel file system.
There are two main options if you lot are working on such a system, which are discussed more in depth in this article by the HDF Group on parallel IO. It can get quite complicated, and the simplest choice is to intelligently split up your dataset into multiple HDF5 files, such that each process can deal with one .h5 file independently of the others.
Documentation
If you Google lmdb, at least in the United Kingdom, the third search result is IMDb, the Internet Movie Database. That'south not what you were looking for!
Really, in that location is one main source of documentation for the Python bounden of LMDB, which is hosted on Read the Docs LMDB. While the Python package hasn't even reached version > 0.94, it is quite widely used and is considered stable.
As for the LMDB technology itself, there is more detailed documentation at the LMDB technology website, which tin experience a flake like learning calculus in 2nd form, unless you showtime from their Getting Started page.
For HDF5, in that location is very clear documentation at the h5py docs site, as well as a helpful weblog post past Christopher Lovell, which is an splendid overview of how to use the h5py parcel. The O'Reilly book, Python and HDF5 besides is a good way to get started.
While not equally documented as possibly a beginner would appreciate, both LMDB and HDF5 accept large user communities, so a deeper Google search usually yields helpful results.
A More Critical Look at Implementation
There is no utopia in storage systems, and both LMDB and HDF5 accept their share of pitfalls.
A key indicate to understand virtually LMDB is that new data is written without overwriting or moving existing data. This is a design decision that allows for the extremely quick reads you lot witnessed in our experiments, and also guarantees data integrity and reliability without the additional need of keeping transaction logs.
Remember, however, that you needed to define the map_size parameter for retentiveness allocation before writing to a new database? This is where LMDB tin can exist a hassle. Suppose you have created an LMDB database, and everything is wonderful. You lot've waited patiently for your enormous dataset to be packed into a LMDB.
Then, afterwards downwardly the line, yous think that y'all demand to add new data. Even with the buffer you specified on your map_size, you may easily expect to see the lmdb.MapFullError fault. Unless you want to re-write your entire database, with the updated map_size, you'll have to shop that new data in a separate LMDB file. Fifty-fifty though one transaction can span multiple LMDB files, having multiple files can still be a pain.
Additionally, some systems accept restrictions on how much memory may be claimed at once. In my ain experience, working with high-performance computing (HPC) systems, this has proved extremely frustrating, and has often made me adopt HDF5 over LMDB.
With both LMDB and HDF5, only the requested item is read into retention at once. With LMDB, key-unit of measurement pairs are read into retention i by one, while with HDF5, the dataset object tin be accessed similar a Python array, with indexing dataset[i], ranges, dataset[i:j] and other splicing dataset[i:j:interval].
Because of the way the systems are optimized, and depending on your operating system, the order in which you access items can impact operation.
In my experience, information technology'southward generally true that for LMDB, you lot may get amend performance when accessing items sequentially by central (fundamental-value pairs being kept in memory ordered alphanumerically past primal), and that for HDF5, accessing large ranges will perform better than reading every element of the dataset i by ane using the following:
# Slightly slower for i in range ( len ( dataset )): # Read the ith value in the dataset, one at a time do_something_with ( dataset [ i ]) # This is improve data = dataset [:] for d in information : do_something_with ( d ) If you are considering a choice of file storage format to write your software around, it would exist remiss not to mention Moving away from HDF5 past Cyrille Rossant on the pitfalls of HDF5, and Konrad Hinsen'south response On HDF5 and the future of information management, which shows how some of the pitfalls tin can exist avoided in his ain utilise cases with many smaller datasets rather than a few enormous ones. Note that a relatively smaller dataset is nevertheless several GB in size.
Integration With Other Libraries
If y'all're dealing with really big datasets, it's highly probable that you'll be doing something significant with them. It's worthwhile to consider deep learning libraries and what kind of integration there is with LMDB and HDF5.
First of all, all libraries support reading images from deejay as .png files, as long as y'all convert them into NumPy arrays of the expected format. This holds true for all the methods, and nosotros have already seen higher up that information technology is relatively straightforward to read in images every bit arrays.
Here are several of the most popular deep learning libraries and their LMDB and HDF5 integration:
-
Caffe has a stable, well-supported LMDB integration, and information technology handles the reading pace transparently. The LMDB layer can likewise hands be replaced with a HDF5 database.
-
Keras uses the HDF5 format to salve and restore models. This implies that TensorFlow can as well.
-
TensorFlow has a born grade
LMDBDatasetthat provides an interface for reading in input information from an LMDB file and can produce iterators and tensors in batches. TensorFlow does not have a congenital-in form for HDF5, but one tin be written that inherits from theDatasetcourse. I personally use a custom class birthday that is designed for optimal read access based on the way I construction my HDF5 files. -
Theano does not natively back up any detail file format or database, but as previously stated, can utilize annihilation as long as it is read in every bit an N-dimensional array.
While far from comprehensive, this hopefully gives you a experience for the LMDB/HDF5 integration by some fundamental deep learning libraries.
A Few Personal Insights on Storing Images in Python
In my own daily work analyzing terabytes of medical images, I utilize both LMDB and HDF5, and have learned that, with whatsoever storage method, forethought is critical.
Often, models demand to be trained using k-fold cross validation, which involves splitting the unabridged dataset into k-sets (g typically being x), and k models being trained, each with a different yard-fix used every bit test set up. This ensures that the model is non overfitting the dataset, or, in other words, unable to make good predictions on unseen data.
A standard way to arts and crafts a 1000-gear up is to put an equal representation of each type of data represented in the dataset in each thou-set. Thus, saving each k-fix into a separate HDF5 dataset maximizes efficiency. Sometimes, a single k-set cannot be loaded into memory at once, so fifty-fifty the ordering of data within a dataset requires some forethought.
With LMDB, I similarly am careful to program ahead before creating the database(due south). There are a few skilful questions worth asking earlier you save images:
- How tin I save the images such that about of the reads will be sequential?
- What are good keys?
- How can I summate a good
map_size, anticipating potential future changes in the dataset? - How big can a single transaction be, and how should transactions be subdivided?
Regardless of the storage method, when you lot're dealing with large image datasets, a little planning goes a long fashion.
Conclusion
You've fabricated it to the end! You've now had a bird'southward eye view of a big topic.
In this article, y'all've been introduced to three ways of storing and accessing lots of images in Python, and possibly had a chance to play with some of them. All the code for this article is in a Jupyter notebook here or Python script here. Run at your own risk, equally a few GB of your disk space volition be overtaken by little foursquare images of cars, boats, and so on.
Yous've seen evidence of how various storage methods can drastically affect read and write time, as well as a few pros and cons of the three methods considered in this commodity. While storing images as .png files may be the most intuitive, at that place are large performance benefits to considering methods such equally HDF5 or LMDB.
Feel free to discuss in the comment section the excellent storage methods not covered in this commodity, such as LevelDB, Feather, TileDB, Badger, BoltDB, or anything else. There is no perfect storage method, and the best method depends on your specific dataset and employ cases.
Further Reading
Here are some references related to the three methods covered in this commodity:
- Python binding for LMDB
- LMDB documentation: Getting Started
- Python bounden for HDF5 (h5py)
- The HDF5 Group
- "Python and HDF5" from O'Reilly
- Pillow
You lot may as well appreciate "An analysis of prototype storage systems for scalable training of deep neural networks" by Lim, Young, and Patton. That paper covers experiments similar to the ones in this article, simply on a much larger calibration, considering common cold and warm enshroud also as other factors.
Source: https://realpython.com/storing-images-in-python/
0 Response to "Reading All Images in a Folder Python"
Post a Comment